Calculate and compare network properties for microbial networks using Jaccard's index, the Rand index, the Graphlet Correlation Distance, and permutation tests.
Usage
netCompare(
x,
permTest = FALSE,
jaccQuant = 0.75,
lnormFit = NULL,
testRand = TRUE,
nPermRand = 1000L,
gcd = TRUE,
gcdOrb = c(0, 2, 5, 7, 8, 10, 11, 6, 9, 4, 1),
verbose = TRUE,
nPerm = 1000L,
adjust = "adaptBH",
trueNullMethod = "convest",
cores = 1L,
logFile = NULL,
seed = NULL,
fileLoadAssoPerm = NULL,
fileLoadCountsPerm = NULL,
storeAssoPerm = FALSE,
fileStoreAssoPerm = "assoPerm",
storeCountsPerm = FALSE,
fileStoreCountsPerm = c("countsPerm1", "countsPerm2"),
returnPermProps = FALSE,
returnPermCentr = FALSE,
assoPerm = NULL,
dissPerm = NULL
)Arguments
- x
object of class
microNetProps(returned bynetAnalyze).- permTest
logical. If
TRUE, a permutation test is conducted to test centrality measures and global network properties for group differences. Defaults toFALSE. May lead to a considerably increased execution time!- jaccQuant
numeric value between 0 and 1 specifying the quantile used as threshold to identify the most central nodes for each centrality measure. The resulting sets of nodes are used to calculate Jaccard's index (see details). Default is 0.75.
- lnormFit
logical indicating whether a log-normal distribution should be fitted to the calculated centrality values for determining Jaccard's index (see details). If
NULL(default), the value is adopted from the input, i.e., equals the method used for determining hub nodes.- testRand
logical. If
TRUE, a permutation test is conducted for the adjusted Rand index (with H0: ARI = 0). Execution time may be increased for large networks.- nPermRand
integer giving the number of permutations used for testing the adjusted Rand index for being significantly different from zero. Ignored if
testRand = FALSE. Defaults to 1000L.- gcd
logical. If
TRUE(default), the Graphlet Correlation Distance (GCD) is computed.- gcdOrb
numeric vector with integers from 0 to 14 defining the orbits used for calculating the GCD. Minimum length is 2. Defaults to c(0, 1, 2, 4, 5, 6, 7, 8, 9, 10, 11), thus excluding redundant orbits such as the orbit o3.
- verbose
logical. If
TRUE(default), status messages are shown.- nPerm
integer giving the number of permutations if
permTest = TRUE. Default is 1000L.- adjust
character indicating the method used for multiple testing adjustment of the permutation p-values. Possible values are
"lfdr"(default) for local false discovery rate correction (viafdrtool),"adaptBH"for the adaptive Benjamini-Hochberg method (Benjamini and Hochberg, 2000), or one of the methods provided byp.adjust(seep.adjust.methods()).- trueNullMethod
character indicating the method used for estimating the proportion of true null hypotheses from a vector of p-values. Used for the adaptive Benjamini-Hochberg method for multiple testing adjustment (chosen by
adjust = "adaptBH"). Accepts the provided options of themethodargument ofpropTrueNull:"convest"(default),"lfdr","mean", and"hist". Can alternatively be"farco"for the "iterative plug-in method" proposed by Farcomeni (2007).- cores
integer indicating the number of CPU cores used for permutation tests. If cores > 1, the tests are performed in parallel. Is limited to the number of available CPU cores determined by
detectCores. Defaults to 1L (no parallelization).- logFile
character string naming the log file to which the current iteration number is written (if permutation tests are performed). Defaults to
NULLso that no log file is generated.- seed
integer giving a seed for reproducibility of the results.
- fileLoadAssoPerm
character giving the name or path (without file extension) of the file containing the "permuted" association/dissimilarity matrices that was generated by setting
storeAssoPermtoTRUE. Only used for permutation tests. IfNULL, no existing associations are used.- fileLoadCountsPerm
character giving the name or path (without file extension) of the file containing the "permuted" count matrices that was generated by setting
storeCountsPermtoTRUE. Only used for permutation tests, and iffileLoadAssoPerm = NULL. IfNULL, no existing count matrices are used.- storeAssoPerm
logical indicating whether the association/dissimilarity matrices for the permuted data should be saved to a file. The file name is given via
fileStoreAssoPerm. IfTRUE, the computed "permutation" association/dissimilarity matrices can be reused viafileLoadAssoPermto save runtime. Defaults toFALSE. Ignored iffileLoadAssoPermis notNULL.- fileStoreAssoPerm
character giving the name of a file to which the matrix with associations/dissimilarities of the permuted data is saved. Can also be a path.
- storeCountsPerm
logical indicating whether the permuted count matrices should be saved to an external file. Defaults to
FALSE. Ignored iffileLoadCountsPermis notNULL.- fileStoreCountsPerm
character vector with two elements giving the names of two files storing the permuted count matrices belonging to the two groups.
- returnPermProps
logical. If
TRUE, the global properties and their absolute differences for the permuted data are returned.- returnPermCentr
logical. If
TRUE, the centralities and their absolute differences for the permuted data are returned.- assoPerm
only needed for output generated with NetCoMi v1.0.1! A list with two elements used for the permutation procedure. Each entry must contain association matrices for
"nPerm"permutations. This can be the"assoPerm"value as part of the output either returned bydiffnetornetCompare.- dissPerm
only needed for output generated with NetCoMi v1.0.1! Usage analog to
assoPermif a dissimilarity measure has been used for network construction.
Value
Object of class microNetComp with the following
elements:
jaccDeg,jaccBetw,jaccClose,jaccEigen | Values of Jaccard's index for the centrality measures |
jaccHub | Jaccard index for the sets of hub nodes |
randInd | Adjusted Rand index |
randIndLCC | Adjusted Rand index for the largest connected component (LCC) |
gcd | Graphlet Correlation Distance (object of class gcd
returned by calcGCD) |
gcdLCC | Graphlet Correlation Distance for the LCC |
properties | List with calculated network properties |
propertiesLCC | List with calculated network properties of the LCC |
diffGlobal | Vectors with differences of global properties |
diffGlobalLCC | Vectors with differences of global properties for the LCC |
diffCent | Vectors with differences of the centrality values |
countMatrices | The two count matrices returned
by netConstruct |
assoMatrices | The two association matrices returned
by netConstruct |
dissMatrices | The two dissimilarity matrices returned
by netConstruct |
adjaMatrices | The two adjacency matrices returned
by netConstruct |
groups | Group names returned by netConstruct |
paramsProperties | Parameters used for network analysis |
Additional output if permutation tests are conducted:
pvalDiffGlobal | P-values of the tests for differential global properties |
pvalDiffGlobalLCC | P-values of the tests for differential global properties in the LCC |
pvalDiffCentr | P-values of the tests for differential centrality values |
pvalDiffCentrAdjust | Adjusted p-values of the tests for differential centrality values |
permDiffGlobal | nPerm x 10 matrix containing the absolute
differences of the ten global network properties (computed for the whole
network) for all nPerm permutations |
permDiffGlobalLCC | nPerm x 11 matrix containing the
absolute differences of the eleven global network properties (computed for
the LCC) for all nPerm permutations |
permDiffCentr | List with absolute differences of the four
centrality measures for all nPerm permutations. Each list contains
a nPerm x nNodes matrix. |
Details
Permutation procedure:
Used for testing centrality measures and global network properties for
group differences.
The null hypothesis of the tests is defined as
$$H_0: c1_i - c2_i = 0,$$ where \(c1_i\) and
\(c2_i\) denote the centrality values of taxon i in group 1 and 2,
respectively.
To generate a sampling distribution of the differences under \(H_0\),
the group labels are randomly reassigned to the samples while the group
sizes are kept. The associations are then re-estimated for each permuted
data set. The p-values are calculated as the proportion of
"permutation-differences" being larger than or equal to the observed
difference. In non-exact tests, a pseudo-count is added to the numerator
and denominator to avoid p-values of zero. Several methods for adjusting
the p-values for multiplicity are available.
Jaccard's index:
Jaccard's index expresses for each centrality measure how equal the sets of
most central nodes are among the two networks.
These sets are defined as nodes with a centrality value above a defined
quantile (via jaccQuant) either of the empirical distribution of the
centrality values (lnormFit = FALSE) or of a fitted log-normal
distribution (lnormFit = TRUE).
The index ranges from 0 to 1, where 1 means the sets of most central nodes
are exactly equal in both networks and 0 indicates that the
most central nodes are completely different.
The index is calculated as suggested by Real and Vargas (1996).
Rand index:
The Rand index is used to express whether the determined clusterings are
equal in both groups. The adjusted Rand index (ARI) ranges from -1 to 1,
where 1 indicates that the two clusterings are exactly equal. The expected
index value for two random clusterings is 0. The implemented test procedure
is in accordance with the explanations in Qannari et al. (2014),
where a p-value below the alpha levels means that ARI is significantly
higher than expected for two random clusterings.
Graphlet Correlation Distance:
A graphlet-based distance measure, which is defined as the Euclidean
distance of the upper triangle values of the Graphlet Correlation
Matrices (GCM) of two networks (Yaveroglu et al., 2014).
The GCM of a network is a matrix with Spearman's correlations between the
network's node orbits (Hocevar and Demsar, 2016).
See calcGCD for details.
References
Benjamini Y, Hochberg Y (2000). “On the adaptive control of the false
discovery rate in multiple testing with independent statistics.”
Journal of Educational and Behavioral Statistics, 25(1), 60–83.
Farcomeni A (2007). “Some results on the control of the false discovery
rate under dependence.” Scandinavian Journal of Statistics,
34(2), 275–297.
Gill R, Datta S, Datta S (2010). “A statistical framework for differential
network analysis from microarray data.” BMC Bioinformatics,
11, 95.
Hocevar T, Demsar J (2016). “Computation of graphlet orbits for nodes and
edges in sparse graphs.” Journal of Statistical Software,
71, 1–24.
Qannari EM, Courcoux P, Faye P (2014). “Significance test of the adjusted
Rand index. Application to the free sorting task.”
Food Quality and Preference, 32, 93–97.
Real R, Vargas JM (1996). “The Probabilistic Basis of Jaccard's Index of
Similarity.” Systematic Biology, 45, 380–385.
Yaveroglu ON, Malod-Dognin N, Davis D, Levnajic Z, Janjic V, Karapandza R,
Stojmirovic A, Przulj N (2014). “Revealing the hidden language of complex
networks.” Scientific reports, 4(1), 1–9.
Examples
knitr::opts_chunk$set(fig.width = 16, fig.height = 8)
# Load data sets from American Gut Project (from SpiecEasi package)
data("amgut2.filt.phy")
# Split data into two groups: with and without seasonal allergies
amgut_season_yes <- phyloseq::subset_samples(amgut2.filt.phy,
SEASONAL_ALLERGIES == "yes")
amgut_season_no <- phyloseq::subset_samples(amgut2.filt.phy,
SEASONAL_ALLERGIES == "no")
amgut_season_yes
#> phyloseq-class experiment-level object
#> otu_table() OTU Table: [ 138 taxa and 121 samples ]
#> sample_data() Sample Data: [ 121 samples by 166 sample variables ]
#> tax_table() Taxonomy Table: [ 138 taxa by 7 taxonomic ranks ]
amgut_season_no
#> phyloseq-class experiment-level object
#> otu_table() OTU Table: [ 138 taxa and 163 samples ]
#> sample_data() Sample Data: [ 163 samples by 166 sample variables ]
#> tax_table() Taxonomy Table: [ 138 taxa by 7 taxonomic ranks ]
# Filter the 121 samples (sample size of the smaller group) with highest
# frequency to make the sample sizes equal and thus ensure comparability.
n_yes <- phyloseq::nsamples(amgut_season_yes)
# Network construction
amgut_net <- netConstruct(data = amgut_season_yes,
data2 = amgut_season_no,
measure = "pearson",
filtSamp = "highestFreq",
filtSampPar = list(highestFreq = n_yes),
filtTax = "highestVar",
filtTaxPar = list(highestVar = 30),
zeroMethod = "pseudoZO", normMethod = "clr")
#> Checking input arguments ...
#> Done.
#> Data filtering ...
#> 0 samples removed in data set 1.
#> 42 samples removed in data set 2.
#> 114 taxa removed in each data set.
#> 1 rows with zero sum removed in group 1.
#> 24 taxa and 120 samples remaining in group 1.
#> 24 taxa and 121 samples remaining in group 2.
#>
#> Zero treatment in group 1:
#> Zero counts replaced by 1
#>
#> Zero treatment in group 2:
#> Zero counts replaced by 1
#>
#> Normalization in group 1:
#> Execute clr(){SpiecEasi} ...
#> Done.
#>
#> Normalization in group 2:
#> Execute clr(){SpiecEasi} ...
#> Done.
#>
#> Calculate 'pearson' associations ...
#> Done.
#>
#> Calculate associations in group 2 ...
#> Done.
#>
#> Sparsify associations via 't-test' ...
#>
#> Adjust for multiple testing via 'adaptBH' ...
#> Done.
#> Done.
#>
#> Sparsify associations in group 2 ...
#>
#> Adjust for multiple testing via 'adaptBH' ...
#> Done.
#> Done.
# Network analysis
# Note: Please zoom into the GCM plot or open a new window using:
# x11(width = 10, height = 10)
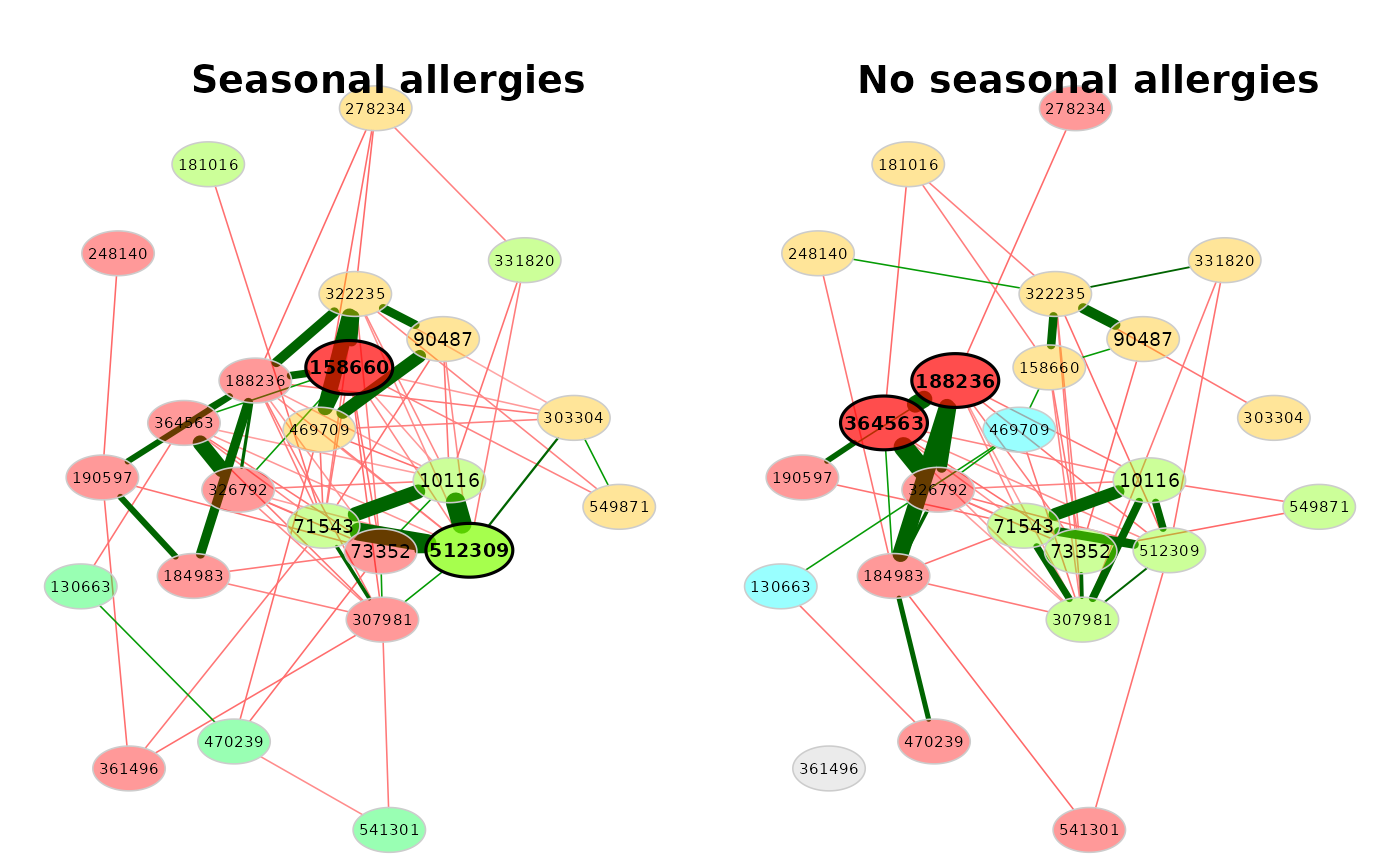
amgut_props <- netAnalyze(amgut_net, clustMethod = "cluster_fast_greedy")
 # Network plot
plot(amgut_props,
sameLayout = TRUE,
title1 = "Seasonal allergies",
title2 = "No seasonal allergies")
# Network plot
plot(amgut_props,
sameLayout = TRUE,
title1 = "Seasonal allergies",
title2 = "No seasonal allergies")
 #--------------------------
# Network comparison
# Without permutation tests
amgut_comp1 <- netCompare(amgut_props, permTest = FALSE)
#> Checking input arguments ...
#> Done.
summary(amgut_comp1)
#>
#> Comparison of Network Properties
#> ----------------------------------
#> CALL:
#> netCompare(x = amgut_props, permTest = FALSE)
#>
#> ______________________________
#> Global network properties
#> `````````````````````````
#> Whole network:
#> group '1' group '2' difference
#> Number of components 1.000 2.000 1.000
#> Clustering coefficient 0.534 0.448 0.086
#> Modularity 0.168 0.155 0.012
#> Positive edge percentage 32.099 39.683 7.584
#> Edge density 0.293 0.249 0.044
#> Natural connectivity 0.070 0.068 0.002
#> Vertex connectivity 1.000 1.000 0.000
#> Edge connectivity 1.000 1.000 0.000
#> Average dissimilarity* 0.920 0.929 0.009
#> Average path length** 1.496 1.558 0.062
#> -----
#> *: Dissimilarity = 1 - edge weight
#> **: Path length = Units with average dissimilarity
#>
#> ______________________________
#> Jaccard index (similarity betw. sets of most central nodes)
#> ```````````````````````````````````````````````````````````
#> Jacc P(<=Jacc) P(>=Jacc)
#> degree 0.167 0.351166 0.912209
#> betweenness centr. 0.333 0.650307 0.622822
#> closeness centr. 0.333 0.650307 0.622822
#> eigenvec. centr. 0.333 0.650307 0.622822
#> hub taxa 0.000 0.197531 1.000000
#> -----
#> Jaccard index in [0,1] (1 indicates perfect agreement)
#>
#> ______________________________
#> Adjusted Rand index (similarity betw. clusterings)
#> ``````````````````````````````````````````````````
#> wholeNet LCC
#> ARI 0.054 0.054
#> p-value 0.409 0.397
#> -----
#> ARI in [-1,1] with ARI=1: perfect agreement betw. clusterings
#> ARI=0: expected for two random clusterings
#> p-value: permutation test (n=1000) with null hypothesis ARI=0
#>
#> ______________________________
#> Graphlet Correlation Distance
#> `````````````````````````````
#> wholeNet LCC
#> GCD 1.203 0.954
#> -----
#> GCD >= 0 (GCD=0 indicates perfect agreement between GCMs)
#>
#> ______________________________
#> Centrality measures
#> - In decreasing order
#> - Computed for the whole network
#> ````````````````````````````````````
#> Degree (normalized):
#> group '1' group '2' abs.diff.
#> 158660 0.522 0.261 0.261
#> 469709 0.391 0.174 0.217
#> 303304 0.261 0.043 0.217
#> 184983 0.174 0.348 0.174
#> 10116 0.478 0.304 0.174
#> 512309 0.565 0.391 0.174
#> 278234 0.174 0.043 0.130
#> 361496 0.130 0.000 0.130
#> 71543 0.522 0.391 0.130
#> 188236 0.565 0.435 0.130
#>
#> Betweenness centrality (normalized):
#> group '1' group '2' abs.diff.
#> 184983 0.000 0.147 0.147
#> 322235 0.087 0.195 0.108
#> 190597 0.099 0.000 0.099
#> 188236 0.225 0.143 0.082
#> 71543 0.123 0.043 0.079
#> 512309 0.083 0.139 0.056
#> 326792 0.000 0.043 0.043
#> 73352 0.055 0.095 0.040
#> 248140 0.000 0.026 0.026
#> 278234 0.020 0.000 0.020
#>
#> Closeness centrality (normalized):
#> group '1' group '2' abs.diff.
#> 361496 0.643 0.000 0.643
#> 303304 0.790 0.510 0.280
#> 158660 1.011 0.812 0.200
#> 248140 0.478 0.675 0.197
#> 469709 0.931 0.772 0.159
#> 278234 0.678 0.539 0.139
#> 184983 0.775 0.909 0.135
#> 512309 1.045 0.912 0.133
#> 181016 0.544 0.665 0.121
#> 10116 0.966 0.850 0.115
#>
#> Eigenvector centrality (normalized):
#> group '1' group '2' abs.diff.
#> 158660 0.971 0.314 0.657
#> 184983 0.319 0.774 0.455
#> 322235 0.857 0.403 0.454
#> 469709 0.695 0.309 0.386
#> 303304 0.397 0.037 0.360
#> 90487 0.483 0.200 0.283
#> 307981 0.682 0.965 0.283
#> 364563 0.716 0.990 0.274
#> 326792 0.707 0.954 0.246
#> 512309 1.000 0.828 0.172
#>
#> _________________________________________________________
#> Significance codes: ***: 0.001, **: 0.01, *: 0.05, .: 0.1
# \donttest{
# With permutation tests (with only 100 permutations to decrease runtime)
amgut_comp2 <- netCompare(amgut_props,
permTest = TRUE,
nPerm = 100L,
cores = 1L,
storeCountsPerm = TRUE,
fileStoreCountsPerm = c("countsPerm1",
"countsPerm2"),
storeAssoPerm = TRUE,
fileStoreAssoPerm = "assoPerm",
seed = 123456)
#> Checking input arguments ...
#> Done.
#> Calculate network properties ...
#> Done.
#> Files 'countsPerm1.bmat, countsPerm1.desc.txt,
#> countsPerm2.bmat, and countsPerm2.desc.txt created.
#> Files 'assoPerm.bmat and assoPerm.desc.txt created.
#> Execute permutation tests ...
#>
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%
#> Done.
#> Calculating p-values ...
#> Done.
#> Adjust for multiple testing using 'adaptBH' ...
#> Done.
# Rerun with a different adjustment method ...
# ... using the stored permutation count matrices
amgut_comp3 <- netCompare(amgut_props, adjust = "BH",
permTest = TRUE, nPerm = 100L,
fileLoadCountsPerm = c("countsPerm1",
"countsPerm2"),
seed = 123456)
#> Checking input arguments ...
#> Done.
#> Calculate network properties ...
#> Done.
#> Execute permutation tests ...
#>
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%
#> Done.
#> Calculating p-values ...
#> Done.
#> Adjust for multiple testing using 'BH' ...
#> Done.
# ... using the stored permutation association matrices
amgut_comp4 <- netCompare(amgut_props, adjust = "BH",
permTest = TRUE, nPerm = 100L,
fileLoadAssoPerm = "assoPerm",
seed = 123456)
#> Checking input arguments ...
#> Done.
#> Calculate network properties ...
#> Done.
#> Execute permutation tests ...
#>
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%
#> Done.
#> Calculating p-values ...
#> Done.
#> Adjust for multiple testing using 'BH' ...
#> Done.
# amgut_comp3 and amgut_comp4 should be equal
all.equal(amgut_comp3$adjaMatrices, amgut_comp4$adjaMatrices)
#> [1] TRUE
all.equal(amgut_comp3$properties, amgut_comp4$properties)
#> [1] TRUE
summary(amgut_comp2)
#>
#> Comparison of Network Properties
#> ----------------------------------
#> CALL:
#> netCompare(x = amgut_props, permTest = TRUE, nPerm = 100, cores = 1,
#> seed = 123456, storeAssoPerm = TRUE, fileStoreAssoPerm = "assoPerm",
#> storeCountsPerm = TRUE, fileStoreCountsPerm = c("countsPerm1",
#> "countsPerm2"))
#>
#> ______________________________
#> Global network properties
#> `````````````````````````
#> Whole network:
#> group '1' group '2' abs.diff. p-value
#> Number of components 1.000 2.000 1.000 0.811881
#> Clustering coefficient 0.534 0.448 0.086 0.435644
#> Modularity 0.168 0.155 0.012 0.881188
#> Positive edge percentage 32.099 39.683 7.584 0.108911
#> Edge density 0.293 0.249 0.044 0.524752
#> Natural connectivity 0.070 0.068 0.002 0.891089
#> Vertex connectivity 1.000 1.000 0.000 1.000000
#> Edge connectivity 1.000 1.000 0.000 1.000000
#> Average dissimilarity* 0.920 0.929 0.009 0.643564
#> Average path length** 1.496 1.558 0.062 0.712871
#> -----
#> p-values: one-tailed test with null hypothesis diff=0
#> *: Dissimilarity = 1 - edge weight
#> **: Path length = Units with average dissimilarity
#>
#> ______________________________
#> Jaccard index (similarity betw. sets of most central nodes)
#> ```````````````````````````````````````````````````````````
#> Jacc P(<=Jacc) P(>=Jacc)
#> degree 0.167 0.351166 0.912209
#> betweenness centr. 0.333 0.650307 0.622822
#> closeness centr. 0.333 0.650307 0.622822
#> eigenvec. centr. 0.333 0.650307 0.622822
#> hub taxa 0.000 0.197531 1.000000
#> -----
#> Jaccard index in [0,1] (1 indicates perfect agreement)
#>
#> ______________________________
#> Adjusted Rand index (similarity betw. clusterings)
#> ``````````````````````````````````````````````````
#> wholeNet LCC
#> ARI 0.054 0.054
#> p-value 0.281 0.413
#> -----
#> ARI in [-1,1] with ARI=1: perfect agreement betw. clusterings
#> ARI=0: expected for two random clusterings
#> p-value: permutation test (n=1000) with null hypothesis ARI=0
#>
#> ______________________________
#> Graphlet Correlation Distance
#> `````````````````````````````
#> wholeNet LCC
#> GCD 1.203000 0.95400
#> p-value 0.762376 0.90099
#> -----
#> GCD >= 0 (GCD=0 indicates perfect agreement between GCMs)
#> p-value: permutation test with null hypothesis GCD=0
#>
#> ______________________________
#> Centrality measures
#> - In decreasing order
#> - Computed for the whole network
#> ````````````````````````````````````
#> Degree (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.522 0.261 0.261 0.984441
#> 469709 0.391 0.174 0.217 0.984441
#> 303304 0.261 0.043 0.217 0.984441
#> 184983 0.174 0.348 0.174 0.984441
#> 10116 0.478 0.304 0.174 0.984441
#> 512309 0.565 0.391 0.174 0.984441
#> 278234 0.174 0.043 0.130 0.984441
#> 361496 0.130 0.000 0.130 0.984441
#> 71543 0.522 0.391 0.130 0.984441
#> 188236 0.565 0.435 0.130 0.984441
#>
#> Betweenness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 184983 0.000 0.147 0.147 0.861386
#> 322235 0.087 0.195 0.108 0.891089
#> 190597 0.099 0.000 0.099 0.861386
#> 188236 0.225 0.143 0.082 0.891089
#> 71543 0.123 0.043 0.079 0.891089
#> 512309 0.083 0.139 0.056 1.000000
#> 326792 0.000 0.043 0.043 0.861386
#> 73352 0.055 0.095 0.040 0.891089
#> 248140 0.000 0.026 0.026 0.861386
#> 278234 0.020 0.000 0.020 0.861386
#>
#> Closeness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 361496 0.643 0.000 0.643 0.344046
#> 303304 0.790 0.510 0.280 0.344046
#> 158660 1.011 0.812 0.200 0.796029
#> 248140 0.478 0.675 0.197 0.796029
#> 469709 0.931 0.772 0.159 0.796029
#> 278234 0.678 0.539 0.139 0.796029
#> 184983 0.775 0.909 0.135 0.796029
#> 512309 1.045 0.912 0.133 0.796029
#> 181016 0.544 0.665 0.121 0.815517
#> 10116 0.966 0.850 0.115 0.796029
#>
#> Eigenvector centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.971 0.314 0.657 0.511232
#> 184983 0.319 0.774 0.455 0.511232
#> 322235 0.857 0.403 0.454 0.511232
#> 469709 0.695 0.309 0.386 0.526269
#> 303304 0.397 0.037 0.360 0.315761
#> 90487 0.483 0.200 0.283 0.631522
#> 307981 0.682 0.965 0.283 0.631522
#> 364563 0.716 0.990 0.274 0.511232
#> 326792 0.707 0.954 0.246 0.511232
#> 512309 1.000 0.828 0.172 0.721740
#>
#> _________________________________________________________
#> Significance codes: ***: 0.001, **: 0.01, *: 0.05, .: 0.1
summary(amgut_comp3)
#>
#> Comparison of Network Properties
#> ----------------------------------
#> CALL:
#> netCompare(x = amgut_props, permTest = TRUE, nPerm = 100, adjust = "BH",
#> seed = 123456, fileLoadCountsPerm = c("countsPerm1", "countsPerm2"))
#>
#> ______________________________
#> Global network properties
#> `````````````````````````
#> Whole network:
#> group '1' group '2' abs.diff. p-value
#> Number of components 1.000 2.000 1.000 0.811881
#> Clustering coefficient 0.534 0.448 0.086 0.435644
#> Modularity 0.168 0.155 0.012 0.881188
#> Positive edge percentage 32.099 39.683 7.584 0.108911
#> Edge density 0.293 0.249 0.044 0.524752
#> Natural connectivity 0.070 0.068 0.002 0.891089
#> Vertex connectivity 1.000 1.000 0.000 1.000000
#> Edge connectivity 1.000 1.000 0.000 1.000000
#> Average dissimilarity* 0.920 0.929 0.009 0.643564
#> Average path length** 1.496 1.558 0.062 0.712871
#> -----
#> p-values: one-tailed test with null hypothesis diff=0
#> *: Dissimilarity = 1 - edge weight
#> **: Path length = Units with average dissimilarity
#>
#> ______________________________
#> Jaccard index (similarity betw. sets of most central nodes)
#> ```````````````````````````````````````````````````````````
#> Jacc P(<=Jacc) P(>=Jacc)
#> degree 0.167 0.351166 0.912209
#> betweenness centr. 0.333 0.650307 0.622822
#> closeness centr. 0.333 0.650307 0.622822
#> eigenvec. centr. 0.333 0.650307 0.622822
#> hub taxa 0.000 0.197531 1.000000
#> -----
#> Jaccard index in [0,1] (1 indicates perfect agreement)
#>
#> ______________________________
#> Adjusted Rand index (similarity betw. clusterings)
#> ``````````````````````````````````````````````````
#> wholeNet LCC
#> ARI 0.054 0.054
#> p-value 0.281 0.413
#> -----
#> ARI in [-1,1] with ARI=1: perfect agreement betw. clusterings
#> ARI=0: expected for two random clusterings
#> p-value: permutation test (n=1000) with null hypothesis ARI=0
#>
#> ______________________________
#> Graphlet Correlation Distance
#> `````````````````````````````
#> wholeNet LCC
#> GCD 1.203000 0.95400
#> p-value 0.762376 0.90099
#> -----
#> GCD >= 0 (GCD=0 indicates perfect agreement between GCMs)
#> p-value: permutation test with null hypothesis GCD=0
#>
#> ______________________________
#> Centrality measures
#> - In decreasing order
#> - Computed for the whole network
#> ````````````````````````````````````
#> Degree (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.522 0.261 0.261 0.984441
#> 469709 0.391 0.174 0.217 0.984441
#> 303304 0.261 0.043 0.217 0.984441
#> 184983 0.174 0.348 0.174 0.984441
#> 10116 0.478 0.304 0.174 0.984441
#> 512309 0.565 0.391 0.174 0.984441
#> 278234 0.174 0.043 0.130 0.984441
#> 361496 0.130 0.000 0.130 0.984441
#> 71543 0.522 0.391 0.130 0.984441
#> 188236 0.565 0.435 0.130 0.984441
#>
#> Betweenness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 184983 0.000 0.147 0.147 0.861386
#> 322235 0.087 0.195 0.108 0.891089
#> 190597 0.099 0.000 0.099 0.861386
#> 188236 0.225 0.143 0.082 0.891089
#> 71543 0.123 0.043 0.079 0.891089
#> 512309 0.083 0.139 0.056 1.000000
#> 326792 0.000 0.043 0.043 0.861386
#> 73352 0.055 0.095 0.040 0.891089
#> 248140 0.000 0.026 0.026 0.861386
#> 278234 0.020 0.000 0.020 0.861386
#>
#> Closeness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 361496 0.643 0.000 0.643 0.356436
#> 303304 0.790 0.510 0.280 0.356436
#> 158660 1.011 0.812 0.200 0.824694
#> 248140 0.478 0.675 0.197 0.824694
#> 469709 0.931 0.772 0.159 0.824694
#> 278234 0.678 0.539 0.139 0.824694
#> 184983 0.775 0.909 0.135 0.824694
#> 512309 1.045 0.912 0.133 0.824694
#> 181016 0.544 0.665 0.121 0.844884
#> 10116 0.966 0.850 0.115 0.824694
#>
#> Eigenvector centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.971 0.314 0.657 0.577086
#> 184983 0.319 0.774 0.455 0.577086
#> 322235 0.857 0.403 0.454 0.577086
#> 469709 0.695 0.309 0.386 0.594059
#> 303304 0.397 0.037 0.360 0.356436
#> 90487 0.483 0.200 0.283 0.712871
#> 307981 0.682 0.965 0.283 0.712871
#> 364563 0.716 0.990 0.274 0.577086
#> 326792 0.707 0.954 0.246 0.577086
#> 512309 1.000 0.828 0.172 0.814710
#>
#> _________________________________________________________
#> Significance codes: ***: 0.001, **: 0.01, *: 0.05, .: 0.1
summary(amgut_comp4)
#>
#> Comparison of Network Properties
#> ----------------------------------
#> CALL:
#> netCompare(x = amgut_props, permTest = TRUE, nPerm = 100, adjust = "BH",
#> seed = 123456, fileLoadAssoPerm = "assoPerm")
#>
#> ______________________________
#> Global network properties
#> `````````````````````````
#> Whole network:
#> group '1' group '2' abs.diff. p-value
#> Number of components 1.000 2.000 1.000 0.811881
#> Clustering coefficient 0.534 0.448 0.086 0.435644
#> Modularity 0.168 0.155 0.012 0.881188
#> Positive edge percentage 32.099 39.683 7.584 0.108911
#> Edge density 0.293 0.249 0.044 0.524752
#> Natural connectivity 0.070 0.068 0.002 0.891089
#> Vertex connectivity 1.000 1.000 0.000 1.000000
#> Edge connectivity 1.000 1.000 0.000 1.000000
#> Average dissimilarity* 0.920 0.929 0.009 0.643564
#> Average path length** 1.496 1.558 0.062 0.712871
#> -----
#> p-values: one-tailed test with null hypothesis diff=0
#> *: Dissimilarity = 1 - edge weight
#> **: Path length = Units with average dissimilarity
#>
#> ______________________________
#> Jaccard index (similarity betw. sets of most central nodes)
#> ```````````````````````````````````````````````````````````
#> Jacc P(<=Jacc) P(>=Jacc)
#> degree 0.167 0.351166 0.912209
#> betweenness centr. 0.333 0.650307 0.622822
#> closeness centr. 0.333 0.650307 0.622822
#> eigenvec. centr. 0.333 0.650307 0.622822
#> hub taxa 0.000 0.197531 1.000000
#> -----
#> Jaccard index in [0,1] (1 indicates perfect agreement)
#>
#> ______________________________
#> Adjusted Rand index (similarity betw. clusterings)
#> ``````````````````````````````````````````````````
#> wholeNet LCC
#> ARI 0.054 0.054
#> p-value 0.281 0.413
#> -----
#> ARI in [-1,1] with ARI=1: perfect agreement betw. clusterings
#> ARI=0: expected for two random clusterings
#> p-value: permutation test (n=1000) with null hypothesis ARI=0
#>
#> ______________________________
#> Graphlet Correlation Distance
#> `````````````````````````````
#> wholeNet LCC
#> GCD 1.203000 0.95400
#> p-value 0.762376 0.90099
#> -----
#> GCD >= 0 (GCD=0 indicates perfect agreement between GCMs)
#> p-value: permutation test with null hypothesis GCD=0
#>
#> ______________________________
#> Centrality measures
#> - In decreasing order
#> - Computed for the whole network
#> ````````````````````````````````````
#> Degree (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.522 0.261 0.261 0.984441
#> 469709 0.391 0.174 0.217 0.984441
#> 303304 0.261 0.043 0.217 0.984441
#> 184983 0.174 0.348 0.174 0.984441
#> 10116 0.478 0.304 0.174 0.984441
#> 512309 0.565 0.391 0.174 0.984441
#> 278234 0.174 0.043 0.130 0.984441
#> 361496 0.130 0.000 0.130 0.984441
#> 71543 0.522 0.391 0.130 0.984441
#> 188236 0.565 0.435 0.130 0.984441
#>
#> Betweenness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 184983 0.000 0.147 0.147 0.861386
#> 322235 0.087 0.195 0.108 0.891089
#> 190597 0.099 0.000 0.099 0.861386
#> 188236 0.225 0.143 0.082 0.891089
#> 71543 0.123 0.043 0.079 0.891089
#> 512309 0.083 0.139 0.056 1.000000
#> 326792 0.000 0.043 0.043 0.861386
#> 73352 0.055 0.095 0.040 0.891089
#> 248140 0.000 0.026 0.026 0.861386
#> 278234 0.020 0.000 0.020 0.861386
#>
#> Closeness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 361496 0.643 0.000 0.643 0.356436
#> 303304 0.790 0.510 0.280 0.356436
#> 158660 1.011 0.812 0.200 0.824694
#> 248140 0.478 0.675 0.197 0.824694
#> 469709 0.931 0.772 0.159 0.824694
#> 278234 0.678 0.539 0.139 0.824694
#> 184983 0.775 0.909 0.135 0.824694
#> 512309 1.045 0.912 0.133 0.824694
#> 181016 0.544 0.665 0.121 0.844884
#> 10116 0.966 0.850 0.115 0.824694
#>
#> Eigenvector centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.971 0.314 0.657 0.577086
#> 184983 0.319 0.774 0.455 0.577086
#> 322235 0.857 0.403 0.454 0.577086
#> 469709 0.695 0.309 0.386 0.594059
#> 303304 0.397 0.037 0.360 0.356436
#> 90487 0.483 0.200 0.283 0.712871
#> 307981 0.682 0.965 0.283 0.712871
#> 364563 0.716 0.990 0.274 0.577086
#> 326792 0.707 0.954 0.246 0.577086
#> 512309 1.000 0.828 0.172 0.814710
#>
#> _________________________________________________________
#> Significance codes: ***: 0.001, **: 0.01, *: 0.05, .: 0.1
#--------------------------
# Use 'createAssoPerm' to create "permuted" count and association matrices
createAssoPerm(amgut_props, nPerm = 100,
computeAsso = TRUE,
fileStoreAssoPerm = "assoPerm",
storeCountsPerm = TRUE,
fileStoreCountsPerm = c("countsPerm1", "countsPerm2"),
append = FALSE, seed = 123456)
#> Create matrix with permuted group labels ...
#> Done.
#> Files 'assoPerm.bmat and assoPerm.desc.txt created.
#> Files 'countsPerm1.bmat, countsPerm1.desc.txt, countsPerm2.bmat, and countsPerm2.desc.txt created.
#> Compute permutation associations ...
#>
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%
#> Done.
amgut_comp5 <- netCompare(amgut_props, permTest = TRUE, nPerm = 100L,
fileLoadAssoPerm = "assoPerm")
#> Checking input arguments ...
#> Done.
#> Calculate network properties ...
#> Done.
#> Execute permutation tests ...
#>
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%
#> Done.
#> Calculating p-values ...
#> Done.
#> Adjust for multiple testing using 'adaptBH' ...
#> Done.
all.equal(amgut_comp3$properties, amgut_comp5$properties)
#> [1] TRUE
summary(amgut_comp5)
#>
#> Comparison of Network Properties
#> ----------------------------------
#> CALL:
#> netCompare(x = amgut_props, permTest = TRUE, nPerm = 100, fileLoadAssoPerm = "assoPerm")
#>
#> ______________________________
#> Global network properties
#> `````````````````````````
#> Whole network:
#> group '1' group '2' abs.diff. p-value
#> Number of components 1.000 2.000 1.000 0.811881
#> Clustering coefficient 0.534 0.448 0.086 0.435644
#> Modularity 0.168 0.155 0.012 0.881188
#> Positive edge percentage 32.099 39.683 7.584 0.108911
#> Edge density 0.293 0.249 0.044 0.524752
#> Natural connectivity 0.070 0.068 0.002 0.891089
#> Vertex connectivity 1.000 1.000 0.000 1.000000
#> Edge connectivity 1.000 1.000 0.000 1.000000
#> Average dissimilarity* 0.920 0.929 0.009 0.643564
#> Average path length** 1.496 1.558 0.062 0.712871
#> -----
#> p-values: one-tailed test with null hypothesis diff=0
#> *: Dissimilarity = 1 - edge weight
#> **: Path length = Units with average dissimilarity
#>
#> ______________________________
#> Jaccard index (similarity betw. sets of most central nodes)
#> ```````````````````````````````````````````````````````````
#> Jacc P(<=Jacc) P(>=Jacc)
#> degree 0.167 0.351166 0.912209
#> betweenness centr. 0.333 0.650307 0.622822
#> closeness centr. 0.333 0.650307 0.622822
#> eigenvec. centr. 0.333 0.650307 0.622822
#> hub taxa 0.000 0.197531 1.000000
#> -----
#> Jaccard index in [0,1] (1 indicates perfect agreement)
#>
#> ______________________________
#> Adjusted Rand index (similarity betw. clusterings)
#> ``````````````````````````````````````````````````
#> wholeNet LCC
#> ARI 0.054 0.054
#> p-value 0.373 0.399
#> -----
#> ARI in [-1,1] with ARI=1: perfect agreement betw. clusterings
#> ARI=0: expected for two random clusterings
#> p-value: permutation test (n=1000) with null hypothesis ARI=0
#>
#> ______________________________
#> Graphlet Correlation Distance
#> `````````````````````````````
#> wholeNet LCC
#> GCD 1.203000 0.95400
#> p-value 0.762376 0.90099
#> -----
#> GCD >= 0 (GCD=0 indicates perfect agreement between GCMs)
#> p-value: permutation test with null hypothesis GCD=0
#>
#> ______________________________
#> Centrality measures
#> - In decreasing order
#> - Computed for the whole network
#> ````````````````````````````````````
#> Degree (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.522 0.261 0.261 0.984441
#> 469709 0.391 0.174 0.217 0.984441
#> 303304 0.261 0.043 0.217 0.984441
#> 184983 0.174 0.348 0.174 0.984441
#> 10116 0.478 0.304 0.174 0.984441
#> 512309 0.565 0.391 0.174 0.984441
#> 278234 0.174 0.043 0.130 0.984441
#> 361496 0.130 0.000 0.130 0.984441
#> 71543 0.522 0.391 0.130 0.984441
#> 188236 0.565 0.435 0.130 0.984441
#>
#> Betweenness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 184983 0.000 0.147 0.147 0.861386
#> 322235 0.087 0.195 0.108 0.891089
#> 190597 0.099 0.000 0.099 0.861386
#> 188236 0.225 0.143 0.082 0.891089
#> 71543 0.123 0.043 0.079 0.891089
#> 512309 0.083 0.139 0.056 1.000000
#> 326792 0.000 0.043 0.043 0.861386
#> 73352 0.055 0.095 0.040 0.891089
#> 248140 0.000 0.026 0.026 0.861386
#> 278234 0.020 0.000 0.020 0.861386
#>
#> Closeness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 361496 0.643 0.000 0.643 0.344046
#> 303304 0.790 0.510 0.280 0.344046
#> 158660 1.011 0.812 0.200 0.796029
#> 248140 0.478 0.675 0.197 0.796029
#> 469709 0.931 0.772 0.159 0.796029
#> 278234 0.678 0.539 0.139 0.796029
#> 184983 0.775 0.909 0.135 0.796029
#> 512309 1.045 0.912 0.133 0.796029
#> 181016 0.544 0.665 0.121 0.815517
#> 10116 0.966 0.850 0.115 0.796029
#>
#> Eigenvector centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.971 0.314 0.657 0.511232
#> 184983 0.319 0.774 0.455 0.511232
#> 322235 0.857 0.403 0.454 0.511232
#> 469709 0.695 0.309 0.386 0.526269
#> 303304 0.397 0.037 0.360 0.315761
#> 90487 0.483 0.200 0.283 0.631522
#> 307981 0.682 0.965 0.283 0.631522
#> 364563 0.716 0.990 0.274 0.511232
#> 326792 0.707 0.954 0.246 0.511232
#> 512309 1.000 0.828 0.172 0.721740
#>
#> _________________________________________________________
#> Significance codes: ***: 0.001, **: 0.01, *: 0.05, .: 0.1
# }
#--------------------------
# Network comparison
# Without permutation tests
amgut_comp1 <- netCompare(amgut_props, permTest = FALSE)
#> Checking input arguments ...
#> Done.
summary(amgut_comp1)
#>
#> Comparison of Network Properties
#> ----------------------------------
#> CALL:
#> netCompare(x = amgut_props, permTest = FALSE)
#>
#> ______________________________
#> Global network properties
#> `````````````````````````
#> Whole network:
#> group '1' group '2' difference
#> Number of components 1.000 2.000 1.000
#> Clustering coefficient 0.534 0.448 0.086
#> Modularity 0.168 0.155 0.012
#> Positive edge percentage 32.099 39.683 7.584
#> Edge density 0.293 0.249 0.044
#> Natural connectivity 0.070 0.068 0.002
#> Vertex connectivity 1.000 1.000 0.000
#> Edge connectivity 1.000 1.000 0.000
#> Average dissimilarity* 0.920 0.929 0.009
#> Average path length** 1.496 1.558 0.062
#> -----
#> *: Dissimilarity = 1 - edge weight
#> **: Path length = Units with average dissimilarity
#>
#> ______________________________
#> Jaccard index (similarity betw. sets of most central nodes)
#> ```````````````````````````````````````````````````````````
#> Jacc P(<=Jacc) P(>=Jacc)
#> degree 0.167 0.351166 0.912209
#> betweenness centr. 0.333 0.650307 0.622822
#> closeness centr. 0.333 0.650307 0.622822
#> eigenvec. centr. 0.333 0.650307 0.622822
#> hub taxa 0.000 0.197531 1.000000
#> -----
#> Jaccard index in [0,1] (1 indicates perfect agreement)
#>
#> ______________________________
#> Adjusted Rand index (similarity betw. clusterings)
#> ``````````````````````````````````````````````````
#> wholeNet LCC
#> ARI 0.054 0.054
#> p-value 0.409 0.397
#> -----
#> ARI in [-1,1] with ARI=1: perfect agreement betw. clusterings
#> ARI=0: expected for two random clusterings
#> p-value: permutation test (n=1000) with null hypothesis ARI=0
#>
#> ______________________________
#> Graphlet Correlation Distance
#> `````````````````````````````
#> wholeNet LCC
#> GCD 1.203 0.954
#> -----
#> GCD >= 0 (GCD=0 indicates perfect agreement between GCMs)
#>
#> ______________________________
#> Centrality measures
#> - In decreasing order
#> - Computed for the whole network
#> ````````````````````````````````````
#> Degree (normalized):
#> group '1' group '2' abs.diff.
#> 158660 0.522 0.261 0.261
#> 469709 0.391 0.174 0.217
#> 303304 0.261 0.043 0.217
#> 184983 0.174 0.348 0.174
#> 10116 0.478 0.304 0.174
#> 512309 0.565 0.391 0.174
#> 278234 0.174 0.043 0.130
#> 361496 0.130 0.000 0.130
#> 71543 0.522 0.391 0.130
#> 188236 0.565 0.435 0.130
#>
#> Betweenness centrality (normalized):
#> group '1' group '2' abs.diff.
#> 184983 0.000 0.147 0.147
#> 322235 0.087 0.195 0.108
#> 190597 0.099 0.000 0.099
#> 188236 0.225 0.143 0.082
#> 71543 0.123 0.043 0.079
#> 512309 0.083 0.139 0.056
#> 326792 0.000 0.043 0.043
#> 73352 0.055 0.095 0.040
#> 248140 0.000 0.026 0.026
#> 278234 0.020 0.000 0.020
#>
#> Closeness centrality (normalized):
#> group '1' group '2' abs.diff.
#> 361496 0.643 0.000 0.643
#> 303304 0.790 0.510 0.280
#> 158660 1.011 0.812 0.200
#> 248140 0.478 0.675 0.197
#> 469709 0.931 0.772 0.159
#> 278234 0.678 0.539 0.139
#> 184983 0.775 0.909 0.135
#> 512309 1.045 0.912 0.133
#> 181016 0.544 0.665 0.121
#> 10116 0.966 0.850 0.115
#>
#> Eigenvector centrality (normalized):
#> group '1' group '2' abs.diff.
#> 158660 0.971 0.314 0.657
#> 184983 0.319 0.774 0.455
#> 322235 0.857 0.403 0.454
#> 469709 0.695 0.309 0.386
#> 303304 0.397 0.037 0.360
#> 90487 0.483 0.200 0.283
#> 307981 0.682 0.965 0.283
#> 364563 0.716 0.990 0.274
#> 326792 0.707 0.954 0.246
#> 512309 1.000 0.828 0.172
#>
#> _________________________________________________________
#> Significance codes: ***: 0.001, **: 0.01, *: 0.05, .: 0.1
# \donttest{
# With permutation tests (with only 100 permutations to decrease runtime)
amgut_comp2 <- netCompare(amgut_props,
permTest = TRUE,
nPerm = 100L,
cores = 1L,
storeCountsPerm = TRUE,
fileStoreCountsPerm = c("countsPerm1",
"countsPerm2"),
storeAssoPerm = TRUE,
fileStoreAssoPerm = "assoPerm",
seed = 123456)
#> Checking input arguments ...
#> Done.
#> Calculate network properties ...
#> Done.
#> Files 'countsPerm1.bmat, countsPerm1.desc.txt,
#> countsPerm2.bmat, and countsPerm2.desc.txt created.
#> Files 'assoPerm.bmat and assoPerm.desc.txt created.
#> Execute permutation tests ...
#>
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%
#> Done.
#> Calculating p-values ...
#> Done.
#> Adjust for multiple testing using 'adaptBH' ...
#> Done.
# Rerun with a different adjustment method ...
# ... using the stored permutation count matrices
amgut_comp3 <- netCompare(amgut_props, adjust = "BH",
permTest = TRUE, nPerm = 100L,
fileLoadCountsPerm = c("countsPerm1",
"countsPerm2"),
seed = 123456)
#> Checking input arguments ...
#> Done.
#> Calculate network properties ...
#> Done.
#> Execute permutation tests ...
#>
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%
#> Done.
#> Calculating p-values ...
#> Done.
#> Adjust for multiple testing using 'BH' ...
#> Done.
# ... using the stored permutation association matrices
amgut_comp4 <- netCompare(amgut_props, adjust = "BH",
permTest = TRUE, nPerm = 100L,
fileLoadAssoPerm = "assoPerm",
seed = 123456)
#> Checking input arguments ...
#> Done.
#> Calculate network properties ...
#> Done.
#> Execute permutation tests ...
#>
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%
#> Done.
#> Calculating p-values ...
#> Done.
#> Adjust for multiple testing using 'BH' ...
#> Done.
# amgut_comp3 and amgut_comp4 should be equal
all.equal(amgut_comp3$adjaMatrices, amgut_comp4$adjaMatrices)
#> [1] TRUE
all.equal(amgut_comp3$properties, amgut_comp4$properties)
#> [1] TRUE
summary(amgut_comp2)
#>
#> Comparison of Network Properties
#> ----------------------------------
#> CALL:
#> netCompare(x = amgut_props, permTest = TRUE, nPerm = 100, cores = 1,
#> seed = 123456, storeAssoPerm = TRUE, fileStoreAssoPerm = "assoPerm",
#> storeCountsPerm = TRUE, fileStoreCountsPerm = c("countsPerm1",
#> "countsPerm2"))
#>
#> ______________________________
#> Global network properties
#> `````````````````````````
#> Whole network:
#> group '1' group '2' abs.diff. p-value
#> Number of components 1.000 2.000 1.000 0.811881
#> Clustering coefficient 0.534 0.448 0.086 0.435644
#> Modularity 0.168 0.155 0.012 0.881188
#> Positive edge percentage 32.099 39.683 7.584 0.108911
#> Edge density 0.293 0.249 0.044 0.524752
#> Natural connectivity 0.070 0.068 0.002 0.891089
#> Vertex connectivity 1.000 1.000 0.000 1.000000
#> Edge connectivity 1.000 1.000 0.000 1.000000
#> Average dissimilarity* 0.920 0.929 0.009 0.643564
#> Average path length** 1.496 1.558 0.062 0.712871
#> -----
#> p-values: one-tailed test with null hypothesis diff=0
#> *: Dissimilarity = 1 - edge weight
#> **: Path length = Units with average dissimilarity
#>
#> ______________________________
#> Jaccard index (similarity betw. sets of most central nodes)
#> ```````````````````````````````````````````````````````````
#> Jacc P(<=Jacc) P(>=Jacc)
#> degree 0.167 0.351166 0.912209
#> betweenness centr. 0.333 0.650307 0.622822
#> closeness centr. 0.333 0.650307 0.622822
#> eigenvec. centr. 0.333 0.650307 0.622822
#> hub taxa 0.000 0.197531 1.000000
#> -----
#> Jaccard index in [0,1] (1 indicates perfect agreement)
#>
#> ______________________________
#> Adjusted Rand index (similarity betw. clusterings)
#> ``````````````````````````````````````````````````
#> wholeNet LCC
#> ARI 0.054 0.054
#> p-value 0.281 0.413
#> -----
#> ARI in [-1,1] with ARI=1: perfect agreement betw. clusterings
#> ARI=0: expected for two random clusterings
#> p-value: permutation test (n=1000) with null hypothesis ARI=0
#>
#> ______________________________
#> Graphlet Correlation Distance
#> `````````````````````````````
#> wholeNet LCC
#> GCD 1.203000 0.95400
#> p-value 0.762376 0.90099
#> -----
#> GCD >= 0 (GCD=0 indicates perfect agreement between GCMs)
#> p-value: permutation test with null hypothesis GCD=0
#>
#> ______________________________
#> Centrality measures
#> - In decreasing order
#> - Computed for the whole network
#> ````````````````````````````````````
#> Degree (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.522 0.261 0.261 0.984441
#> 469709 0.391 0.174 0.217 0.984441
#> 303304 0.261 0.043 0.217 0.984441
#> 184983 0.174 0.348 0.174 0.984441
#> 10116 0.478 0.304 0.174 0.984441
#> 512309 0.565 0.391 0.174 0.984441
#> 278234 0.174 0.043 0.130 0.984441
#> 361496 0.130 0.000 0.130 0.984441
#> 71543 0.522 0.391 0.130 0.984441
#> 188236 0.565 0.435 0.130 0.984441
#>
#> Betweenness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 184983 0.000 0.147 0.147 0.861386
#> 322235 0.087 0.195 0.108 0.891089
#> 190597 0.099 0.000 0.099 0.861386
#> 188236 0.225 0.143 0.082 0.891089
#> 71543 0.123 0.043 0.079 0.891089
#> 512309 0.083 0.139 0.056 1.000000
#> 326792 0.000 0.043 0.043 0.861386
#> 73352 0.055 0.095 0.040 0.891089
#> 248140 0.000 0.026 0.026 0.861386
#> 278234 0.020 0.000 0.020 0.861386
#>
#> Closeness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 361496 0.643 0.000 0.643 0.344046
#> 303304 0.790 0.510 0.280 0.344046
#> 158660 1.011 0.812 0.200 0.796029
#> 248140 0.478 0.675 0.197 0.796029
#> 469709 0.931 0.772 0.159 0.796029
#> 278234 0.678 0.539 0.139 0.796029
#> 184983 0.775 0.909 0.135 0.796029
#> 512309 1.045 0.912 0.133 0.796029
#> 181016 0.544 0.665 0.121 0.815517
#> 10116 0.966 0.850 0.115 0.796029
#>
#> Eigenvector centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.971 0.314 0.657 0.511232
#> 184983 0.319 0.774 0.455 0.511232
#> 322235 0.857 0.403 0.454 0.511232
#> 469709 0.695 0.309 0.386 0.526269
#> 303304 0.397 0.037 0.360 0.315761
#> 90487 0.483 0.200 0.283 0.631522
#> 307981 0.682 0.965 0.283 0.631522
#> 364563 0.716 0.990 0.274 0.511232
#> 326792 0.707 0.954 0.246 0.511232
#> 512309 1.000 0.828 0.172 0.721740
#>
#> _________________________________________________________
#> Significance codes: ***: 0.001, **: 0.01, *: 0.05, .: 0.1
summary(amgut_comp3)
#>
#> Comparison of Network Properties
#> ----------------------------------
#> CALL:
#> netCompare(x = amgut_props, permTest = TRUE, nPerm = 100, adjust = "BH",
#> seed = 123456, fileLoadCountsPerm = c("countsPerm1", "countsPerm2"))
#>
#> ______________________________
#> Global network properties
#> `````````````````````````
#> Whole network:
#> group '1' group '2' abs.diff. p-value
#> Number of components 1.000 2.000 1.000 0.811881
#> Clustering coefficient 0.534 0.448 0.086 0.435644
#> Modularity 0.168 0.155 0.012 0.881188
#> Positive edge percentage 32.099 39.683 7.584 0.108911
#> Edge density 0.293 0.249 0.044 0.524752
#> Natural connectivity 0.070 0.068 0.002 0.891089
#> Vertex connectivity 1.000 1.000 0.000 1.000000
#> Edge connectivity 1.000 1.000 0.000 1.000000
#> Average dissimilarity* 0.920 0.929 0.009 0.643564
#> Average path length** 1.496 1.558 0.062 0.712871
#> -----
#> p-values: one-tailed test with null hypothesis diff=0
#> *: Dissimilarity = 1 - edge weight
#> **: Path length = Units with average dissimilarity
#>
#> ______________________________
#> Jaccard index (similarity betw. sets of most central nodes)
#> ```````````````````````````````````````````````````````````
#> Jacc P(<=Jacc) P(>=Jacc)
#> degree 0.167 0.351166 0.912209
#> betweenness centr. 0.333 0.650307 0.622822
#> closeness centr. 0.333 0.650307 0.622822
#> eigenvec. centr. 0.333 0.650307 0.622822
#> hub taxa 0.000 0.197531 1.000000
#> -----
#> Jaccard index in [0,1] (1 indicates perfect agreement)
#>
#> ______________________________
#> Adjusted Rand index (similarity betw. clusterings)
#> ``````````````````````````````````````````````````
#> wholeNet LCC
#> ARI 0.054 0.054
#> p-value 0.281 0.413
#> -----
#> ARI in [-1,1] with ARI=1: perfect agreement betw. clusterings
#> ARI=0: expected for two random clusterings
#> p-value: permutation test (n=1000) with null hypothesis ARI=0
#>
#> ______________________________
#> Graphlet Correlation Distance
#> `````````````````````````````
#> wholeNet LCC
#> GCD 1.203000 0.95400
#> p-value 0.762376 0.90099
#> -----
#> GCD >= 0 (GCD=0 indicates perfect agreement between GCMs)
#> p-value: permutation test with null hypothesis GCD=0
#>
#> ______________________________
#> Centrality measures
#> - In decreasing order
#> - Computed for the whole network
#> ````````````````````````````````````
#> Degree (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.522 0.261 0.261 0.984441
#> 469709 0.391 0.174 0.217 0.984441
#> 303304 0.261 0.043 0.217 0.984441
#> 184983 0.174 0.348 0.174 0.984441
#> 10116 0.478 0.304 0.174 0.984441
#> 512309 0.565 0.391 0.174 0.984441
#> 278234 0.174 0.043 0.130 0.984441
#> 361496 0.130 0.000 0.130 0.984441
#> 71543 0.522 0.391 0.130 0.984441
#> 188236 0.565 0.435 0.130 0.984441
#>
#> Betweenness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 184983 0.000 0.147 0.147 0.861386
#> 322235 0.087 0.195 0.108 0.891089
#> 190597 0.099 0.000 0.099 0.861386
#> 188236 0.225 0.143 0.082 0.891089
#> 71543 0.123 0.043 0.079 0.891089
#> 512309 0.083 0.139 0.056 1.000000
#> 326792 0.000 0.043 0.043 0.861386
#> 73352 0.055 0.095 0.040 0.891089
#> 248140 0.000 0.026 0.026 0.861386
#> 278234 0.020 0.000 0.020 0.861386
#>
#> Closeness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 361496 0.643 0.000 0.643 0.356436
#> 303304 0.790 0.510 0.280 0.356436
#> 158660 1.011 0.812 0.200 0.824694
#> 248140 0.478 0.675 0.197 0.824694
#> 469709 0.931 0.772 0.159 0.824694
#> 278234 0.678 0.539 0.139 0.824694
#> 184983 0.775 0.909 0.135 0.824694
#> 512309 1.045 0.912 0.133 0.824694
#> 181016 0.544 0.665 0.121 0.844884
#> 10116 0.966 0.850 0.115 0.824694
#>
#> Eigenvector centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.971 0.314 0.657 0.577086
#> 184983 0.319 0.774 0.455 0.577086
#> 322235 0.857 0.403 0.454 0.577086
#> 469709 0.695 0.309 0.386 0.594059
#> 303304 0.397 0.037 0.360 0.356436
#> 90487 0.483 0.200 0.283 0.712871
#> 307981 0.682 0.965 0.283 0.712871
#> 364563 0.716 0.990 0.274 0.577086
#> 326792 0.707 0.954 0.246 0.577086
#> 512309 1.000 0.828 0.172 0.814710
#>
#> _________________________________________________________
#> Significance codes: ***: 0.001, **: 0.01, *: 0.05, .: 0.1
summary(amgut_comp4)
#>
#> Comparison of Network Properties
#> ----------------------------------
#> CALL:
#> netCompare(x = amgut_props, permTest = TRUE, nPerm = 100, adjust = "BH",
#> seed = 123456, fileLoadAssoPerm = "assoPerm")
#>
#> ______________________________
#> Global network properties
#> `````````````````````````
#> Whole network:
#> group '1' group '2' abs.diff. p-value
#> Number of components 1.000 2.000 1.000 0.811881
#> Clustering coefficient 0.534 0.448 0.086 0.435644
#> Modularity 0.168 0.155 0.012 0.881188
#> Positive edge percentage 32.099 39.683 7.584 0.108911
#> Edge density 0.293 0.249 0.044 0.524752
#> Natural connectivity 0.070 0.068 0.002 0.891089
#> Vertex connectivity 1.000 1.000 0.000 1.000000
#> Edge connectivity 1.000 1.000 0.000 1.000000
#> Average dissimilarity* 0.920 0.929 0.009 0.643564
#> Average path length** 1.496 1.558 0.062 0.712871
#> -----
#> p-values: one-tailed test with null hypothesis diff=0
#> *: Dissimilarity = 1 - edge weight
#> **: Path length = Units with average dissimilarity
#>
#> ______________________________
#> Jaccard index (similarity betw. sets of most central nodes)
#> ```````````````````````````````````````````````````````````
#> Jacc P(<=Jacc) P(>=Jacc)
#> degree 0.167 0.351166 0.912209
#> betweenness centr. 0.333 0.650307 0.622822
#> closeness centr. 0.333 0.650307 0.622822
#> eigenvec. centr. 0.333 0.650307 0.622822
#> hub taxa 0.000 0.197531 1.000000
#> -----
#> Jaccard index in [0,1] (1 indicates perfect agreement)
#>
#> ______________________________
#> Adjusted Rand index (similarity betw. clusterings)
#> ``````````````````````````````````````````````````
#> wholeNet LCC
#> ARI 0.054 0.054
#> p-value 0.281 0.413
#> -----
#> ARI in [-1,1] with ARI=1: perfect agreement betw. clusterings
#> ARI=0: expected for two random clusterings
#> p-value: permutation test (n=1000) with null hypothesis ARI=0
#>
#> ______________________________
#> Graphlet Correlation Distance
#> `````````````````````````````
#> wholeNet LCC
#> GCD 1.203000 0.95400
#> p-value 0.762376 0.90099
#> -----
#> GCD >= 0 (GCD=0 indicates perfect agreement between GCMs)
#> p-value: permutation test with null hypothesis GCD=0
#>
#> ______________________________
#> Centrality measures
#> - In decreasing order
#> - Computed for the whole network
#> ````````````````````````````````````
#> Degree (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.522 0.261 0.261 0.984441
#> 469709 0.391 0.174 0.217 0.984441
#> 303304 0.261 0.043 0.217 0.984441
#> 184983 0.174 0.348 0.174 0.984441
#> 10116 0.478 0.304 0.174 0.984441
#> 512309 0.565 0.391 0.174 0.984441
#> 278234 0.174 0.043 0.130 0.984441
#> 361496 0.130 0.000 0.130 0.984441
#> 71543 0.522 0.391 0.130 0.984441
#> 188236 0.565 0.435 0.130 0.984441
#>
#> Betweenness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 184983 0.000 0.147 0.147 0.861386
#> 322235 0.087 0.195 0.108 0.891089
#> 190597 0.099 0.000 0.099 0.861386
#> 188236 0.225 0.143 0.082 0.891089
#> 71543 0.123 0.043 0.079 0.891089
#> 512309 0.083 0.139 0.056 1.000000
#> 326792 0.000 0.043 0.043 0.861386
#> 73352 0.055 0.095 0.040 0.891089
#> 248140 0.000 0.026 0.026 0.861386
#> 278234 0.020 0.000 0.020 0.861386
#>
#> Closeness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 361496 0.643 0.000 0.643 0.356436
#> 303304 0.790 0.510 0.280 0.356436
#> 158660 1.011 0.812 0.200 0.824694
#> 248140 0.478 0.675 0.197 0.824694
#> 469709 0.931 0.772 0.159 0.824694
#> 278234 0.678 0.539 0.139 0.824694
#> 184983 0.775 0.909 0.135 0.824694
#> 512309 1.045 0.912 0.133 0.824694
#> 181016 0.544 0.665 0.121 0.844884
#> 10116 0.966 0.850 0.115 0.824694
#>
#> Eigenvector centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.971 0.314 0.657 0.577086
#> 184983 0.319 0.774 0.455 0.577086
#> 322235 0.857 0.403 0.454 0.577086
#> 469709 0.695 0.309 0.386 0.594059
#> 303304 0.397 0.037 0.360 0.356436
#> 90487 0.483 0.200 0.283 0.712871
#> 307981 0.682 0.965 0.283 0.712871
#> 364563 0.716 0.990 0.274 0.577086
#> 326792 0.707 0.954 0.246 0.577086
#> 512309 1.000 0.828 0.172 0.814710
#>
#> _________________________________________________________
#> Significance codes: ***: 0.001, **: 0.01, *: 0.05, .: 0.1
#--------------------------
# Use 'createAssoPerm' to create "permuted" count and association matrices
createAssoPerm(amgut_props, nPerm = 100,
computeAsso = TRUE,
fileStoreAssoPerm = "assoPerm",
storeCountsPerm = TRUE,
fileStoreCountsPerm = c("countsPerm1", "countsPerm2"),
append = FALSE, seed = 123456)
#> Create matrix with permuted group labels ...
#> Done.
#> Files 'assoPerm.bmat and assoPerm.desc.txt created.
#> Files 'countsPerm1.bmat, countsPerm1.desc.txt, countsPerm2.bmat, and countsPerm2.desc.txt created.
#> Compute permutation associations ...
#>
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%
#> Done.
amgut_comp5 <- netCompare(amgut_props, permTest = TRUE, nPerm = 100L,
fileLoadAssoPerm = "assoPerm")
#> Checking input arguments ...
#> Done.
#> Calculate network properties ...
#> Done.
#> Execute permutation tests ...
#>
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%
#> Done.
#> Calculating p-values ...
#> Done.
#> Adjust for multiple testing using 'adaptBH' ...
#> Done.
all.equal(amgut_comp3$properties, amgut_comp5$properties)
#> [1] TRUE
summary(amgut_comp5)
#>
#> Comparison of Network Properties
#> ----------------------------------
#> CALL:
#> netCompare(x = amgut_props, permTest = TRUE, nPerm = 100, fileLoadAssoPerm = "assoPerm")
#>
#> ______________________________
#> Global network properties
#> `````````````````````````
#> Whole network:
#> group '1' group '2' abs.diff. p-value
#> Number of components 1.000 2.000 1.000 0.811881
#> Clustering coefficient 0.534 0.448 0.086 0.435644
#> Modularity 0.168 0.155 0.012 0.881188
#> Positive edge percentage 32.099 39.683 7.584 0.108911
#> Edge density 0.293 0.249 0.044 0.524752
#> Natural connectivity 0.070 0.068 0.002 0.891089
#> Vertex connectivity 1.000 1.000 0.000 1.000000
#> Edge connectivity 1.000 1.000 0.000 1.000000
#> Average dissimilarity* 0.920 0.929 0.009 0.643564
#> Average path length** 1.496 1.558 0.062 0.712871
#> -----
#> p-values: one-tailed test with null hypothesis diff=0
#> *: Dissimilarity = 1 - edge weight
#> **: Path length = Units with average dissimilarity
#>
#> ______________________________
#> Jaccard index (similarity betw. sets of most central nodes)
#> ```````````````````````````````````````````````````````````
#> Jacc P(<=Jacc) P(>=Jacc)
#> degree 0.167 0.351166 0.912209
#> betweenness centr. 0.333 0.650307 0.622822
#> closeness centr. 0.333 0.650307 0.622822
#> eigenvec. centr. 0.333 0.650307 0.622822
#> hub taxa 0.000 0.197531 1.000000
#> -----
#> Jaccard index in [0,1] (1 indicates perfect agreement)
#>
#> ______________________________
#> Adjusted Rand index (similarity betw. clusterings)
#> ``````````````````````````````````````````````````
#> wholeNet LCC
#> ARI 0.054 0.054
#> p-value 0.373 0.399
#> -----
#> ARI in [-1,1] with ARI=1: perfect agreement betw. clusterings
#> ARI=0: expected for two random clusterings
#> p-value: permutation test (n=1000) with null hypothesis ARI=0
#>
#> ______________________________
#> Graphlet Correlation Distance
#> `````````````````````````````
#> wholeNet LCC
#> GCD 1.203000 0.95400
#> p-value 0.762376 0.90099
#> -----
#> GCD >= 0 (GCD=0 indicates perfect agreement between GCMs)
#> p-value: permutation test with null hypothesis GCD=0
#>
#> ______________________________
#> Centrality measures
#> - In decreasing order
#> - Computed for the whole network
#> ````````````````````````````````````
#> Degree (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.522 0.261 0.261 0.984441
#> 469709 0.391 0.174 0.217 0.984441
#> 303304 0.261 0.043 0.217 0.984441
#> 184983 0.174 0.348 0.174 0.984441
#> 10116 0.478 0.304 0.174 0.984441
#> 512309 0.565 0.391 0.174 0.984441
#> 278234 0.174 0.043 0.130 0.984441
#> 361496 0.130 0.000 0.130 0.984441
#> 71543 0.522 0.391 0.130 0.984441
#> 188236 0.565 0.435 0.130 0.984441
#>
#> Betweenness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 184983 0.000 0.147 0.147 0.861386
#> 322235 0.087 0.195 0.108 0.891089
#> 190597 0.099 0.000 0.099 0.861386
#> 188236 0.225 0.143 0.082 0.891089
#> 71543 0.123 0.043 0.079 0.891089
#> 512309 0.083 0.139 0.056 1.000000
#> 326792 0.000 0.043 0.043 0.861386
#> 73352 0.055 0.095 0.040 0.891089
#> 248140 0.000 0.026 0.026 0.861386
#> 278234 0.020 0.000 0.020 0.861386
#>
#> Closeness centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 361496 0.643 0.000 0.643 0.344046
#> 303304 0.790 0.510 0.280 0.344046
#> 158660 1.011 0.812 0.200 0.796029
#> 248140 0.478 0.675 0.197 0.796029
#> 469709 0.931 0.772 0.159 0.796029
#> 278234 0.678 0.539 0.139 0.796029
#> 184983 0.775 0.909 0.135 0.796029
#> 512309 1.045 0.912 0.133 0.796029
#> 181016 0.544 0.665 0.121 0.815517
#> 10116 0.966 0.850 0.115 0.796029
#>
#> Eigenvector centrality (normalized):
#> group '1' group '2' abs.diff. adj.p-value
#> 158660 0.971 0.314 0.657 0.511232
#> 184983 0.319 0.774 0.455 0.511232
#> 322235 0.857 0.403 0.454 0.511232
#> 469709 0.695 0.309 0.386 0.526269
#> 303304 0.397 0.037 0.360 0.315761
#> 90487 0.483 0.200 0.283 0.631522
#> 307981 0.682 0.965 0.283 0.631522
#> 364563 0.716 0.990 0.274 0.511232
#> 326792 0.707 0.954 0.246 0.511232
#> 512309 1.000 0.828 0.172 0.721740
#>
#> _________________________________________________________
#> Significance codes: ***: 0.001, **: 0.01, *: 0.05, .: 0.1
# }
