This demonstrates how to use the NetCoMi workflow when you already have an association matrix on which to base the network.
The QMP data set provided by the SPRING package is used
to demonstrate how NetCoMi is used to analyze a precomputed network
(given as association matrix).
The data set contains quantitative count data (true absolute values),
which SPRING can deal with. See ?QMP for details.
nlambda and rep.num are set to 10 for a
decreased execution time, but should be higher for real data.
library(SPRING)
# Load the QMP data set
data("QMP")
# Run SPRING for association estimation
fit_spring <- SPRING(QMP,
quantitative = TRUE,
lambdaseq = "data-specific",
nlambda = 10,
rep.num = 10,
seed = 123456,
ncores = 1,
Rmethod = "approx",
verbose = FALSE)
# Optimal lambda
opt.K <- fit_spring$output$stars$opt.index
# Association matrix
assoMat <- as.matrix(SpiecEasi::symBeta(fit_spring$output$est$beta[[opt.K]],
mode = "ave"))
rownames(assoMat) <- colnames(assoMat) <- colnames(QMP)The association matrix is now passed to netConstruct to
start the usual NetCoMi workflow. Note that the dataType
argument must be set appropriately.
# Network construction and analysis
net_asso <- netConstruct(data = assoMat,
dataType = "condDependence",
sparsMethod = "none",
verbose = 0)
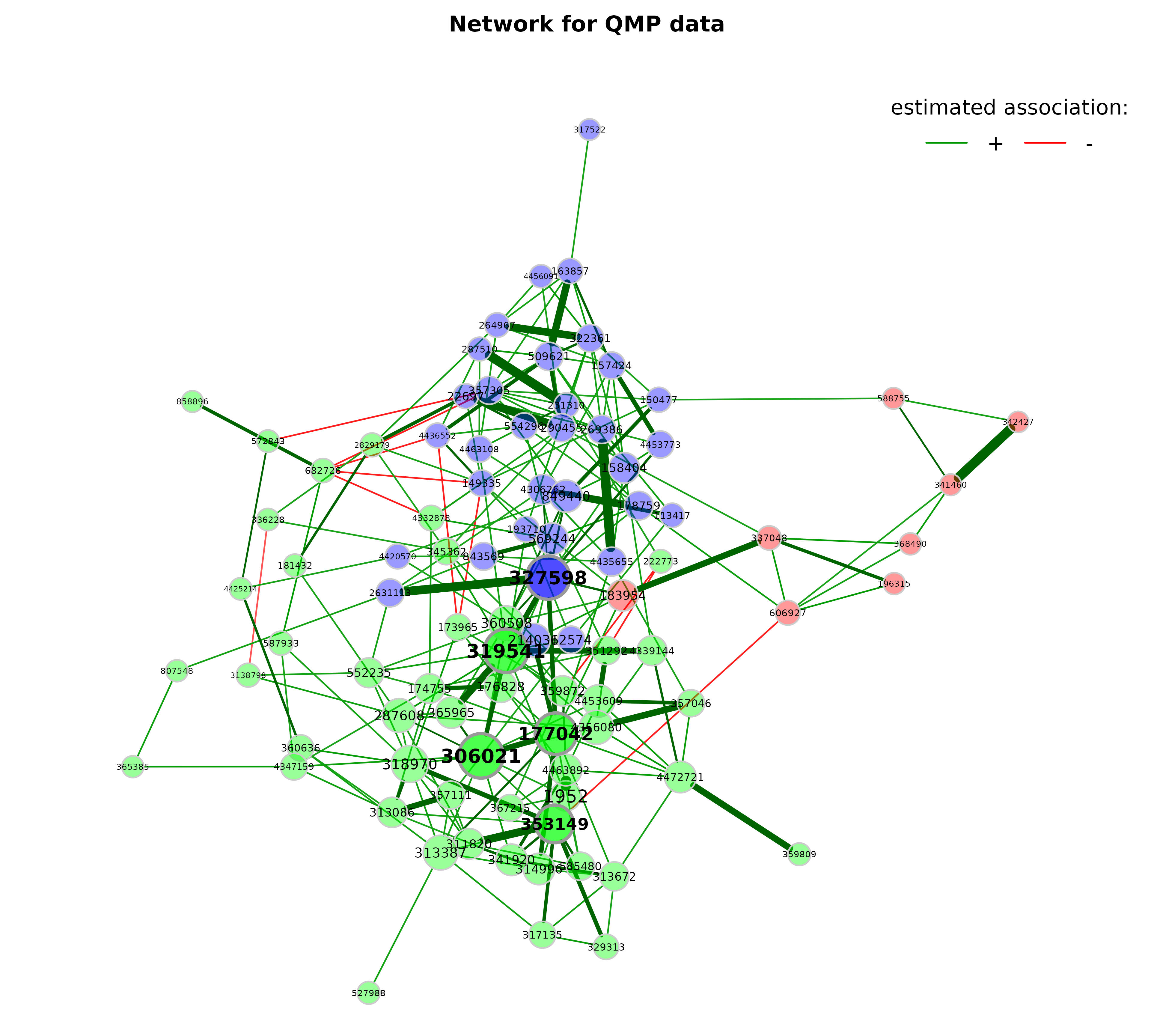
props_asso <- netAnalyze(net_asso, clustMethod = "hierarchical")
plot(props_asso,
layout = "spring",
repulsion = 1.2,
shortenLabels = "none",
labelScale = TRUE,
rmSingles = TRUE,
nodeSize = "eigenvector",
nodeSizeSpread = 2,
nodeColor = "cluster",
hubBorderCol = "gray60",
cexNodes = 1.8,
cexLabels = 2,
cexHubLabels = 2.2,
title1 = "Network for QMP data",
showTitle = TRUE,
cexTitle = 2.3)
legend(0.7, 1.1, cex = 2.2, title = "estimated association:",
legend = c("+","-"), lty = 1, lwd = 3, col = c("#009900","red"),
bty = "n", horiz = TRUE)